In-memory database REDIS provides a simple interface for caching various data types. In addition, it supports simple manipulations with arrays and scalar values, including support for complex data constructs (like queues). What is, however not implemented, is the support of multi-dimensional objects. These objects are beneficial if you need to cache frequently used constants (or interim results) for computations (like matrices). There, however, exist a simple way to circumvent this obstacle.
Using Pandas DataFrame with REDIS
DataFrame in Pandas library represents a popular way how to store 2D structures in Python. It provides a useful encapsulation of NumPy n-dimensional arrays that supports things like labels for columns and rows and SQL-like operations with DataFrame objects. However, it also has well-known problems, mainly related to low performance and frequent changes in API. But despite its problems, Pandas DataFrame popularity is unquestionable. To create a DataFrame object, you first need to install Pandas (pip install pandas), then use the following logic:
import pandas as pd
df = pd.DataFrame(data=NUMPY_ARRAY,
columns=[col1, col2, ...],
index=[row1,row2,...])where NUMPY_ARRAY is a 2D array in NumPy (or just a simple plain Python list that is internally converted to NumPy array), columns argument defines column labels, and index argument defines row labels. For storing multi-dimensional data (more than 2D), Pandas has a simple construct called MultiIndex. It is, however, good to notice that using Pandas for storing more than 2D data is not a common approach (and there are better tools to deal with this problem). Consider the following (simple) example:
df = pd.DataFrame(
data=NUMPY_ARRAY,
columns=[col1, col2, ...],
index=[[row1,row2,...], [idx1, idx2,...]]
)Data here are accessible using a column key and a set of 2-row keys (technically 2D key on the row space). In this way, it is possible to use as many dimensions as required. However, the more sophisticated way to index data is to use MultiIndex class directly, which offers a lot of possibilities (that are not discussed here, but you might find them helpful for some exceptional cases).
Performance issue
The critical question is how to access stored data quickly, primarily when repeatedly accessed in crucial infrastructure. In order to do so, there is a simple way to store the content of the DataFrame using serialization technology called MessagePack (which is now not directly supported in Pandas but can be used externally). MessagePack presents an interesting way that combines binary serialization with plain-text serialization (like JSON or XML). Technically, it is a binary represented JSON object. Another way is to use Pickle, a standard text serialization (native in Pandas and still fully supported). It is worth noting that MessagePack is roughly ten times faster than using Pickle. Another issue with Pickle is that it does not support variable output streams.
Using REDIS in-memory database
A typical way where to store (cache) the serialized object is on the local drive. The disadvantage of this approach in high-performance applications is evident - it is always slower to read from disk than from operational memory (throughput is lower, access time is much higher). On the other hand, it is much more expensive to cache in memory. To avoid delays, you can use REDIS as the cache for serialized DataFrames.
For storing the data, use the following logic:
import redis
import pandas as pd
# ... code
# Connect to redis:
connector = redis.StrictRedis(host="?",
port=?,
password="?")
connector.set("key-in-redis", dataframe.to_msgpack())For reading the data, use the following logic:
import redis
import pandas as pd
# After connecting to the REDIS (described above):
data_frame = connector.get("key-in-redis")These examples illustrate the most straightforward way of how to store data in REDIS and accessing them. It is possible to use every other method in REDIS to work with the binary stream from MessagePack (aka MSG).
Performance analysis
We performed basic measurements of the time required to fetch data from memory and deserialize them to the DataFrame object. The processor Intel Xeon CPU E5-2673 v4 @ 2.30 GHz has been used during measurement. Data are depicted in the following table:
| Number of entities | Time [s] | Standard deviation of time [s] |
|---|---|---|
| 100,000 | 0.003 | 0.002 |
| 500,000 | 0.016 | 0.005 |
| 1,000,000 | 0.034 | 0.012 |
| 5,000,000 | 0.369 | 0.037 |
| 10,000,000 | 0.860 | 0.021 |
| 50,000,000 | 4.324 | 0.041 |
The first column in this table represents the number of entities in a DataFrame (is equal to the number of rows multiplied by the number of columns). The second column is the most important one - it represents the time needed to fetch and deserialize values from REDIS back to the DataFrame. Finally, the third column is the standard deviation of the measured values. In each case, precisely ten measurements are done. Data are also presented in the figure below.
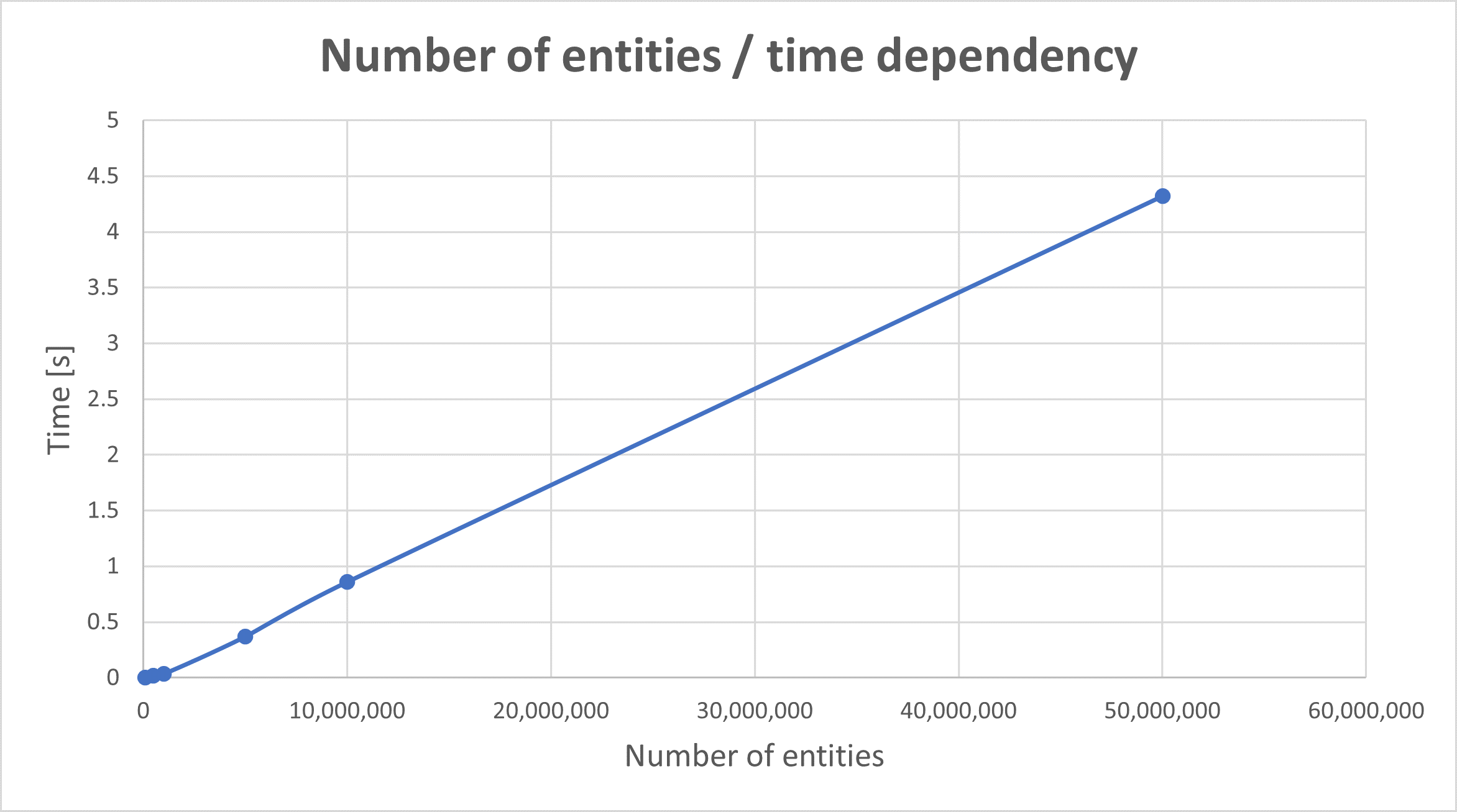
In the figure above, you can see that the time dependency is almost perfectly linear. Also, there is no significant delay up to one million entities (meaning the dealy more than one second). One million entities (table cells) are also an edge value for the optimal size of the stored table (regarding both the price for storing and the performance). Furthermore, it means that a user experience is not affected by the long time needed for receiving values below the edge value. Of course, this value is deeply dependent on the hardware (system) you use; therefore, it is essential to measure each specific hardware's threshold. Also, one second can be too long for particular applications (the acceptable latency should always be defined in requirements). It is also worth to notice, that measurement does not include any delay (latency) caused by network connection (which should not be omitted in practical cases).
Other ways for binary data serialization
All other ways for binary serialization of data are naturally acceptable input for REDIS - if you can use the stream directly. The most popular (and helpful) tools for binary serialization are Apache Avro, gRPC and Apache Thrift. All these RPC frameworks contain parts dedicated to binary data serialization and deserialization. Naturally, the serialization and deserialization process related to each of these technologies requires its overhead. Also, most of the underpinning Interface Description Languages (IDLs) does not support multi-dimensional arrays - so it has to be converted to a 1D array with additional parameters describing dimension lengths.
The general rules and use-case for REDIS
The biggest drawback of the presented example is the time needed for serialization and deserialization. However, this can be overcome if you use some native formats - like n-dimensional symmetric numeric arrays - which supports native serialization (to REDIS). Although REDIS does not support n-dimensional arrays, you can overcome this issue by mapping it to a 1D array (this mapping is trivial as the physical representation in memory is always a one-dimension array). For example, NumPy supports a simple method to perform this operation (flattening the n-dimensional input).
If it comes to particular cases - say geospatial data - be aware of the potentially massive size of these data sets. It can be a big hurdle in successfully speeding up your system using the in-memory database approach. Technically, you can achieve slightly bigger performance for huge prices (or no better performance if you choose the wrong way for the serialization process). Therefore, it is necessary to examine the requirements for your system thoroughly. For example, if you want to share your data among many workers in a message-passing pattern, you need to be aware of latencies when accessing the REDIS instance from the worker.
Many other aspects are critical if you need to increase the performance of your system. The most notorious example is the dimension order of multi-dimensional data. There is a whole theory behind this. Also, in a cloud environment, where the REDIS instance runs on a different machine than your primary application, delays caused by the latencies in communication can be critical.
It is also still true that when dealing with any database (including REDIS), optimising your queries in advance is beneficial. For example, if possible, always read data in one call rather than in many separate calls as decreasing database hits significantly increases overall performance. This approach holds for all database technologies and is well known in the world of relational databases.
Different approaches for caching
The main advantage of caching into the REDIS instance is that your values can be shared but come with costs related to latencies when accessing REDIS. If you need to cache locally, there is a simpler way to do so - the most straightforward way is to use the lru_cache decorator from the functools package (there is a separate article about caching here). There are also many ready-made tools for caching in various frameworks - like Django (that internally uses mainly REDIS, sometimes Memcached).
Sometimes, if your application is correctly optimized, even reading data from a disk can be sufficient. This is the most common case when you process an extended data set - you usually need to load all auxiliary data once at the beginning. As the process is relatively slow, it does not matter a lot if it takes a second or five. Also, when using fast SSD disks, you can reach a comparable performance for in-memory caching. The price is generally much lower when dealing with disk space - so this line of reasoning should be reflected in the design phase.
Conclusions and further research
This article proves that storing a multi-dimensional array of values using a REDIS in-memory database (when dealing with Pandas DataFrame in Python) is suitable for accelerating the system's overall performance. Furthermore, it shows that the cutting value of the table size is one million elements for Intel Xeon CPU E5-2673 v4 @ 2.30 GHz processor. We also discuss other possibilities for caching and serialization of multi-dimensional (and scalar) data like in-place caching using decorators or fast SSD. Finally, some general rules for reading from databases are reminded.
❋ Tags: Pandas Big Data Performance Geospatial REDIS